Welcome
Overview
Teaching: 15 min
Exercises: 0 minQuestions
Who are we and what are we going to learn?
Objectives
Introduce ourselves, and the course
Setup our learning platform
Better rearch by better sharing
Introductions
Introductions set the stage for learning.
— Tracy Teal, Former Executive Director, The Carpentries
Hello everyone, and welcome to FAIR in biological practice workshop. We are very pleased to have you with us.
Today’s Trainers
To begin class, each Trainer should give a brief introduction of themselves.
Now we would like to get to know all of you.
Who are you and what are your expectations from the workshop
Please introduce yourself shortly and tell us:
- Why are you taking this course?
- What goals do you have for the follwing days?
Better research by better sharing
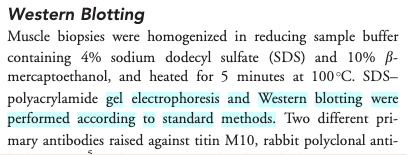
For many of us, data management or outpust sharing in general are considered a burden rather than a useful activity. Part of the problem is our bad timing and lack of planing.
Data management is a continuous process
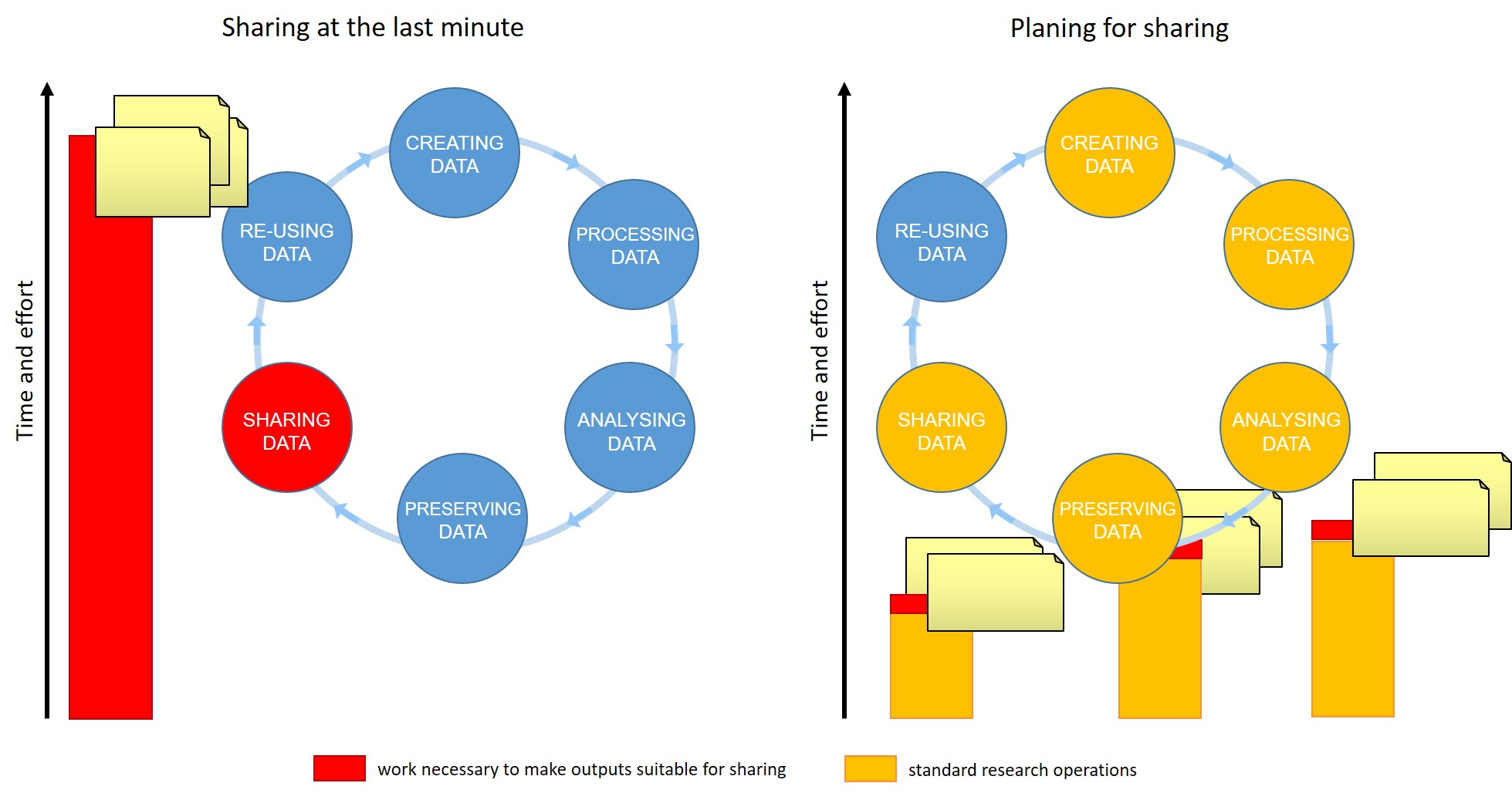 Figure credits: Tomasz Zielinski and Andrés Romanowski
Figure credits: Tomasz Zielinski and Andrés Romanowski
When should you engage in data sharing and open practices?
- Data management should be done throughout the duration of your project.
- If you wait till the end, it will take a massive effort on your side and will be more of a burden than a benefit.
- Taking the time to do effective data management will help you understand your data better and make it easier to find it when you need it (for example when you need to write a manuscript or a thesis!).
- All the practices that enable others accessing and using your outcomes directly benefit you and your group
In this workshop we will show you how you can plan and do your research so in a way that it makes your outputs readily available for re-use by others.
Our agenda:
- Day 1 We will start with explaining Open Science principles and what are the benefits of being open for you and the whole society. Then we will talk about FAIR principles which define steps you we shout take that our “shared” outputs are of value. We will finish with how to plan our work to deliver FAIR outputs.
- Day 2 We will show the benefits of using online records for documenting experiments. We will talk about working and organizing files and using Excel or CSV tables to store and document data.
- Day 3 We will teach how to describe your projects using simple text files or customized templates. We will talk about Version Control.
- Day 4 We will show you how public repositories makes your outputs accessible and reusable. We will consolidate our knowledge of FAIR ready data managment and what other tools can help you during your research.
Online workshop specifics
Our learning tools
Before we begin let’s explain how to use the tools:
- Raising hands,
- Yes/No sticker
- Chatroom for links not for jokes
- Breakout rooms, leaving and rejoining
- using pad, answering questions in pad
- where to find things If needed, check the pre workshop setup, ask to report problems and help at a break or after the session.
Key Points
Do not be shy
Be nice
Remember you can do better research by planing for sharing
Introduction to Open Science
Overview
Teaching: 25 min
Exercises: 25 minQuestions
What is Open Science?
How can I benefit from Open Science?
Why has Open Science become a hot topic?
Objectives
Identify parts of the Open Science movement, their goals and motivations
Explain the main benefits of Open Science
Recognize the barriers and risks in the adoption of Open Science practices
Science works best by exchanging ideas and building on them. Most efficient science involves both questions and experiments being made as fully informed as possible, which requires the free exchange of data and information.
All practices that make knowledge and data freely available fall under the umbrella-term of Open Science/Open Research. It makes science more reproducible, transparent, and accessible. As science becomes more open, the way we conduct and communicate science changes continuously.
What is the Open Science
Open science is the movement to make scientific research (including publications, data, physical samples, and software) and its dissemination accessible to all levels of an inquiring society, amateur or professional.
Open Science represents a new approach to the scientific process based on cooperative work and new ways of diffusing knowledge by using digital technologies and new collaborative tools
Open science is transparent and accessible knowledge that is shared and developed through collaborative networks.
Characteristics:
- Using web-based tools to facilitate information exchange and scientific collaboration
- Transparency in experimental methodology, observation, and collection of data
- Public availability and reusability of scientific data, methods and communications
What is the Open Science movement?
Sharing of information is fundamental for science. This began at a significant scale with the invention of scientific journals in 1665. At that time this was the best available alternative to critique & disseminate research, and foster communities of like-minded researchers.
Whilst this was a great step forward, the journal-driven system of science has led to a culture of ‘closed’ science, where knowledge or data is unavailable or unaffordable to many.
The distribution of knowledge has always been subject to improvement. Whilst the internet was initially developed for military purposes, it was hijacked for communication between scientists, which provided a viable route to change the dissemination of science.
The momentum has built up with a change in the way science is communicated to reflect what research communities are calling for – solutions to the majority of problems (e.g. impact factors, data reusability, reproducibility crisis, trust in the public science sector etc…) that we face today.
Open Science is the movement to increase transparency and reproducibility of research, through using the open best practices.
After Gema Bueno de la Fuente
Open Science Building Blocks
-
Open Access: Research outputs hosted in a way that make them accessible for everyone. Traditionally Open Access referred to journal articles, but now includes books, chapters or images.
-
Open Data: Data freely and readily available to access, reuse, and share. Smaller data sets were often accessible as supplemental materials by journals alongside articles themselves. However, they should be hosted in dedicated platforms for more convenient and better access.
-
Open Software: Software where the source code is made readily available; others are free to use, change, and share. Some examples of these including the coding language and supporting software R and RStudio, as well as image analysis software such as Fiji/ImageJ.
-
Open Notebooks: Lab & notebooks hosted online, readily accessible to all. These are popular among some of the large funding bodies and allow anyone to comment on any stage of the experimental record.
-
Open Peer Review: A system where peer review reports are published alongside the body of work. This can include reviewers’ reports, correspondence between parties involved, rebuttals, editorial decisions etc…
-
Citizen science: Citizen participation of various stages of research process from project funding to collecting and analysing data.
Outcomes/Advantages of Open Science (4+6)
Being open has other outcomes/consequences beyond giving the free access to information. For example, Open educational resources:
- enables collaborative development of courses
- improves teachers/instructors skills by sharing ideas
Select one or two of the following OS parts:
- Open Access
- Open Data
- Open Software
- Open Notebooks
- Open Peer Review
and discuss what are the outcomes or what problems are solved by adaption of those Open initiatives.
Solution
Possible outcomes and consequences for each part:
Open Access
- speed of knowledge distribution
- leveling field for underfunded sites which otherwise wouldn’t be able to navigate the paywall
- prevent articles being paid for ‘thrice’ (first to produce, second to publish, third to access) by institutions.
- greater access to work by others, increasing chance for exposure & citations
- access to work by lay audiences, thus increases social exposure of research
Open Data
- ensures data isn’t lost overtime
- acceleration of scientific discovery rate
- permits statistical re-analysis of the data to validate findings
- gives access to datasets which were not published as papers (e.g. negative results, large screening data sets)
- provides an avenue to generate new hypotheses
- permits combination of multiple data sources to address questions, provides greater power than a single data source
Open Software
- great source to learn programming skills
- the ability to modify creates a supportive community of users and rapid innovation
- faster bug fixes
- better error scrutiny
- use of the same software/code allows better reproducibility between experiments
Open Notebooks
- 100% transparent science, allowing input from others at early stages of experiments
- source of learning about the process of how science is actually conducted
- allows access to experiments and data which otherwise never get published
- provides access to ‘negative’ results and failed experiments
- anyone, anywhere around the world, at any time, can check in on projects, including many users simultaneously
- thorough evidence of originality of ideas and experiments, negating effect of ‘scooping’
Open Peer Review
- visibility leads to more constructive reviews
- mitigates against editorial conflicts of interest and/or biases
- mitigates against reviewers conflicts of interest and/or biases
- allows readers to learn/benefit from comments of the reviewers
Motivation: Money
One has to consider the moral objectives that accompany the research/publication process: charities/taxpayers pay to fund research, these then pay again to access the research they already funded.
From an economic point of view, scientific outputs generated by public research are a public good that everyone should be able to use at no cost.
According to EU report “Cost-benefit analysis for FAIR research data”, €10.2bn is lost every year cause of not accessible data (plus addition 16bn if accounting for re-use and research quality).
The goals of Open Science is to make research and research data available to e.g. charities/taxpayers who funded this research.
COAlition S, a group of national research funding organisations backed by the European Commission and the European Research Council, is a big driver trying to get rid of the paywalls that our research is sat behind. They announced Plan S, an initiative to make research publications fully free at the point of access, meaning that all research funded by public funding bodies must be published Open Access from 2021 onwards.
Open Access (a successful example)
The majority of larger UK and other countries’ funding bodies are now making Open Access publication conditional upon funding.
The initiative is known as Plan S, which requires “resulting publications available immediately (without embargoes) and under open licences, either in quality Open Access platforms or journals or through immediate deposit in open repositories that fulfil the necessary conditions.”
Exact requirements differ between funding bodies, with the minimum requirement being that a copy be deposited with your home institution.
Details of funding bodies and their involvement and requirements can be found at Plan S/cOAlition S. There is also a cOAlition S journal checker tool to assess compliance being developed. The Directory of Open Access Journals (DOAJ) is a tool to find which journals are Open Access.
Motivation: Reproducibility
The inherited transparency of Open Science and the easy access to data, methods and analysis details naturally help to address the Reproducibility crisis. The openness of scientific communications and of the actual process of evaluation of the research (Open Peer Review) increase confidence in the research findings.
Personal motivators
Open Science is advantageous to many parties involved in science (including researcher community, funding bodies, the public even journals), which is leading to a push for the widespread adoption of Open Science practices.
Large UK funding bodies such as The Wellcome Trust are big supporters of Open Science. We can see with the example of Open Access, that once enforced by funders (the stick) there is a wide adoption. But what about the personal motivators, the carrots.
Personal benefits of being “open” (3+3)
Below are some personal benefits to adopting Open Science practices. Read through them which of them are the strongest motivators for you. Select two the most important/attractive for you and mark them with +1, select the two least important for you and mark them with 0
- receive higher citations
- comply with funders’ policies
- get extra value from your work (e.g. collaborators, reuse by modelers, ML specialists)
- demonstrate research impact
- save own time (reproducibility but also communication overhead)
- distinguish yourself from the crowd
- plan successful research proposals
- gain valuable experience
- form community
- increased speed and ease of writing papers
- speed up and help with peer review
- build reputation and presence in the science community
- evidence of your scientific rigour and work ethic
Can you think of other benefits? How personal benefits of Open Science compare to the benefits for the (scientific) society?
The main difference between the public benefits of Open Science practices and the personal motivators of outputs creators, that the public can benefit almost instantly from the open resources. However, the advantages for data creator comes with a delay, typically counted in years. For example, building reputation will not happen with one dataset, the re-use also will lead to citations/collaboration after the next research cycle.
Barriers and risks of OS movement:
Why we are not doing Open Science already (3+3)
Discuss why you did not or would not make your data or software open.
Solution
- sensitive data
- IP
- misuse (fake news)
- lack of confidence (the fear of critics)
- the costs in $ and in time
It may seem obvious that we should adopt open science practices, but there are associated challenges with doing so.
Sensitivity of data is sometimes considered a barrier. Shared data needs to be compliant with data privacy laws, leading many to shy away from hosting it publicly. Anonymising data to desensitise it can help overcome this barrier.
The potential for intellectual property on research can dissuade some from adopting open practices. Again, much can be shared if the data is filtered carefully to protect anything relating to intellectual property.
Open Science and Intellectual property
This section is tailored to the EU and UK.
Intellectual property (IP) is something that you create using your mind - for example, a story, an invention, an artistic work or a symbol.
The timeline of “openning” matters when one seeks legal protection for his IP.
For example, patents are granted only for inventions that are new and were not known to the public in any form. Publishing in a journal or presenting in a conference information related to the invention completely prevents the inventor from getting a patent!
In our opinion, you are more likely to benefit from new collaborations, industrial partnerships, consultations which are acquired by openness, than from patent related royalties.
(Optional) Intellectual property protection
This section is tailored to the EU and UK.
You can use a patent to protect your (technical) invention. It gives you the right to take legal action against anyone who makes, uses, sells or imports it without your permission.
Discoveries, mathematical methods, computer programs and business methods as such are not regarded as inventions. Surgical and therapeutic procedures, diagnostic methods and new plant or animal varieties are completely excluded from patentability.
Patents are granted only for inventions that are new and were not known to the public in any form. Publishing in a journal or presenting in a conference information related to the invention completely prevents the inventor from getting a patent!
In principle, software cannot be patented. However, it is a “no but yes” situation, and a software patent are being granted. It is usually, settled by the court for each case.
Software code is copyrighted. Copyright prevents people from:
- copying your code
- distributing copies of it, whether free of charge or for sale.
Data cannot be patented, and in principle, it cannot be copyrighted. It is not possible to copyright facts!
However, how data are collated and presented (especially if it is a database), can have a layer of copyright protection. Deciding what data needs to be included in a database, how to organize the data, and how to relate different data elements are all creative decisions that may receive copyright protection. Again, it is often a case by case situation and may come down to who has better lawyers.
After: UK Government, Intellectual Property European Patent Office, Inventor’s Handbook
Another risk could be seen with work on Covid19: pre-prints. A manuscript hosted publicly prior to peer review, may accelerate access to knowledge, but can also be misused and/or misunderstood. This can result in political and health decision making based on faulty data, which is counter to societies’ best interest.
One concern is that opening up ones data to the scientific community can lead to the identification of errors, which may lead to feelings of embarrassment. However, this could be considered an upside - we should seek for our work to be scrutinized and errors to be pointed out, and is the sign of a competent scientist. One should rather have errors pointed out rather than risking that irreproducible data might cause even more embarrassment and disaster.
One of the biggest barriers are the costs involved in “being Open”. Firstly, making outputs readily available and usable to others takes time and significant effort. Secondly, there are costs of hosting and storage. For example, microscopy datasets reach sizes in terabytes, making such data accessible for 10 years involves serious financial commitment.
Get involved
Thankfully, incentive structures are beginning to support Open Science practices:
- Universities signing up to the Declaration on Research Assessment (DORA).
- Wellcome Trust funding proposals that increase Open Science
- Wellcome Trust asked for description of Open Science activities in the grant application
You do not want to be left behind!
Where to next
Attribution
Content of this episode was adopted after:
Open Science Quiz (2 + runs over break)
Which of the following statements about the OS movement are true/false?
- Open Science relies strongly on the Internet
- Open Access eliminates publishing costs
- You cannot Open Source patented software
- You cannot earn money from Open Source software
- Open Data facilitates re-use
- Open Data can increases confidence in research findings
- In Open Peer Review, readers vote on publication acceptance
- Open Notebooks improve reproducibility
- Open Notebooks can create patenting issues
- Open Access permits the whole society to benefit from scientific findings
Solution
- Open Science relies strongly on the Internet T
- Open Access eliminates publishing costs F
- You cannot Open Source patented software F*
- You cannot earn money Open Source software F
- Open Data facilitates re-use T
- Open Data increases confidence in research findings T
- In Open Peer Review, readers vote on publication acceptance F
- Open Notebooks improve reproducibility T
- Open Notebooks can create patenting issues T*
- Open Access permits the whole society to benefit from scientific findings T
** For patenting, the dates of public release and patent application are important. Once patent is granted all the details can be made public. On the other hand, laboratory notes published before patent application can be treated as public disclosure and prevents from obtaining a patent.
Key Points
Open Science increases transparency in research
Publicly funded science should be publicly available
Being FAIR
Overview
Teaching: 25 min
Exercises: 25 minQuestions
How to get more value from your own data?
What are the FAIR guidelines?
Why being FAIR matters?
Objectives
Recognize typical issues that prevent data re-use
Understand FAIR principles
Know steps for achieving FAIR data
We have seen how Open practices can benefit both scientific community as a whole and individual practitioner. The wide adoption of Open Access principles has resulted in an easy access to the recent biomedical publications. Unfortunately, the same cannot be said about data and software that accompanies those publications.
What is data
Although scientific data is a very broad term, we still encounter groups who (wrongly) believe they do not have data! Data does not mean Excel files with recorded measurements from a machine. Data also includes:
- images, not only from microscopes
- information about biological materials, like strain or patient details
- recipes, laboratory and measurement protocols
- scripts, analysis procedures, and custom software can also be considered data However, there are specific recommendations on how to deal with code.
Let’s have a look how challenging it can be to access and use data from published biological papers.
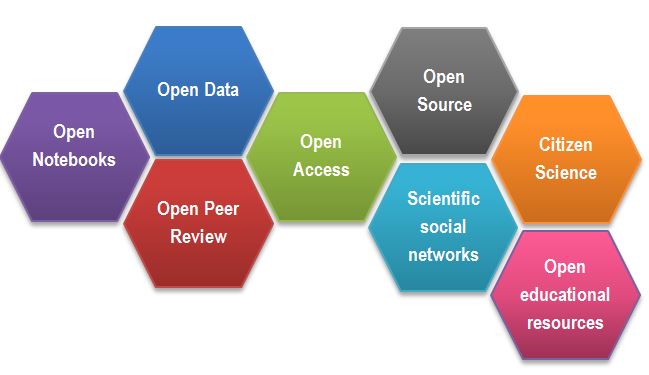
Impossible protocol (5+3)
You need to do a western blot to identify Titin proteins, the largest proteins in the body, with a molecular weight of 3,800 kDa. You found an antibody sold by Sigma Aldrich that has been validated in western blots and immunofluorescence. Sigma Aldrich lists the Yu et al., 2019 paper as reference.
Find details of how to separate and transfer this large protein in the reference paper.
Hint 1: Methods section has a Western blot analysis subsection. Hint 2: Follow the references.
Would you say that the methods was Findable? Accessible? Reusable?
Solution
- Ref 17 will lead you to this paper, which first of all is not Open Access
- Access the paper through your institutions (if you can) and find the ‘Western Blotting’ protocol on page 232 which will show the following (Screenshot from the methods section from Evilä et al 2014):
- “Western blotting were performed according to standard methods.” - with no further reference to these standard methods, describing these methods, or supplementary material detailing these methods
- This methodology is unfortunately a true dead end and we thus can’t easily continue our experiments!
Impossible numbers
Systems biologists usually require raw numerical data to build their models. Take a look at the following example: Try to find the numerical data behind the graph shown in Figure 6 which demonstrates changes in levels of phytochrome proteins from Sharrock RA and Clack T, 2002.
Hint 1: Materials and methods describe quantification procedure Hint 2: Supporting Information or Supplementary Materials sections often contain data files.
How easy it was?
Impossible resource/link
RNA-seq (transcriptomics) data is usually deposited in online repositories such as SRA or ArrayExpress. Your task is to find the link to the repository of the raw RNA-seq data in Li et al., Genes Dev. 2012. Can you find it anywhere?
Impossible format
Sometimes raw data is shared in a proprietary format that is not easily accessible by everyone. Check the format in which xxx is shared in xxxx and compare it to the list of recommended formats. Does the resource comply with recommended guidelines?
The above examples illustrate the typical challenges in accessing research data and software. Firstly, data/protocols/software often do not have an identity of their own, but only accompany a publication. Second, they are not easily accessible or reusable, for example, all the details are inside one supporting information PDF file. Such file includes “printed” numerical table or even source code, both of which need to be “re-typed” if someone would like to use them. Data are shared in proprietary file format specific to a particular vendor and not accessible if one does not have a particular software that accompanies the equipment. Finally, data files are provided without detailed description other than the whole article text.
In our examples, the protocol was difficult to find (the loops), difficult to access (pay wall), and not reusable as it lacked the necessary details (dead-end). In the second example the data were not interoperable and reusable as their were only available as a figure graph.
To avoid such problems FAIR principles were designed.
 After SangyaPundir
After SangyaPundir
FAIR Principles
In 2016, the FAIR Guiding Principles for scientific data management and stewardship were published in Scientific Data. The original guideline focused on “machine-actionability” - the ability of computer systems to operate on data with minimal human intervention. However, now the focus has shifted to making data accessible from a human perspective, and not an automated one (mostly due to the lack of user friendly tools that could help deal with standards and structured metadata).
Findable: Easy to find data and metadata for both humans and computers. Automatic and reliable discovery of datasets and services depends on machine-readable persistent identifiers (PIDs) and metadata.
Accessible: (Meta)data should be retrievable by their identifier using a standardized and open communications protocol (including authentication and authorisation). Metadata should be available even when the data are no longer available.
Interoperable: Data should be able to be combined with and used with other data or tools. The format of the data should be open and interpretable for various tools. It applies both to data and metadata, (meta)data should use vocabularies that follow FAIR principles.
Re-usable: FAIR aims at optimizing the reuse of data. Metadata and data should be well-described so that they can be replicated and/or combined in different settings. The reuse of (meta)data should be stated with clear and accessible license(s).
FAIR in biological practice
Findable & Accessible
Deposit data to an external, reputable public repository.
Repositories provide persistent identifiers (PIDs), catalogue options, advanced metadata searching, and download statistics. Some repositories can also host private data or provide embargo periods, meaning access to all data can be delayed.
There are general “data agnostic” repositories, for example: Dryad, Zenodo, FigShare, Dataverse. Or domain specific, for example: UniProt protein data, GenBank sequence data, MetaboLights metabolomics data and GitHub for code.
We will cover repositories in more details in a later episode.
What are persistent identifiers (PIDs)
A persistent identifier is a long-lasting reference to a digital resource. Typically it has two components:
- a service that locates the resource over time even when its location changes
- and a unique identifier (that distinguishes the resource or concept from others).
Persistent identifiers aim to solve the problem of the persistence of accessing cited resource, particularly in the field of academic literature. All too often, web addresses (links) changes over time and fail to take you to the referenced resource you expected.
There are several services and technologies (schemes) that provide PIDs for objects (whether digital, physical or abstract). One of the most popular is Digital Object Identifier (DOI), recognizable by the prefix doi.org in the web links. For example: https://doi.org/10.1038/sdata.2016.18 resolves to the location of the paper that describes FAIR principles.
Public repositories often maintain web addresses of their content in a stable form which follow the convention http://repository.adress/identifier; these are often called permalinks. For well establish services, permalinks can be treated as PIDs.
For example: http://identifiers.org/SO:0000167 resolves to a page defining promoter role, and can be used to annotate part of a DNA sequence as performing such a role during transcription.
Interoperable
- Use common file formats (can be domain specific)
- Always use .csv or .xls files for numerical data. Never share data tables as word or pdf,
- Provide underlying numerical data for all plots and graphs
- Convert proprietary binary formats to the open ones. For example convert Snapgene to Genbank, microscopy multistack images to OME-TIFF
Reusable
- Describe your data well / provide good metadata
- write README file describing the data
- user descriptive column headers for the data tables
- tidy data tables, make them analysis friendly
- provide as many details as possible (prepare good metadata)
- use (meta)data formats (e.g. SBML, SBOL)
- follow Minimum Information Standards
Describing data well is the most challenging part of the data sharing process. We will cover this in more detail later on.
- Attach license files.
Licenses explicitly declare conditions and terms by which data and software can be re-used.
Here, we recommend:
- for data Creative Commons Attribution (CC BY) license,
- for code a permissive open source license such as the MIT, BSD, or Apache license.
Copyright and data
Software code (the text) automatically gets the default copyright protection which prevents others from copying or modifying it. Only by adding the explicit licence you can permit re-use by others.
Data, being factual, cannot be copyrighted. So why, do we need a licence?
While the data itself cannot be copyrighted, the way how it is presented can be. The extend to which it is protected needs ultimately to be settled by the court.
The “good actors” will restrain from using your data to avoid “court” risks. The “bad actors” will either ignore the risk or can afford the lawyers fees.
Achieving FAIR (2)
Which part of Findable, Accessible, Interoperable, Reusable seems to be the easiest to achieve by you and which looks like the most challenging for data producers.
Solution
Accessible is probably the easiest to achieve as it can be delivered by depositing to any of the general repositories. Findable as accessible by persistent identifiers (PIDs) is also easy as offered by the same repositories. However, findable in the sense of search results by some features and characteristics may not be so simple.
Interoperable and Reusable are the most challenging as they require substantial effort and time to convert existing data into well documented and in suitable format.
Example of FAIR data (5+3)
The Nucleotide database is a collection of sequences from several sources, including GenBank, RefSeq, TPA and PDB. Genome, gene and transcript sequence data provide the foundation for biomedical research and discovery.
Have a look at record for myoglobin gen https://www.ncbi.nlm.nih.gov/nuccore/AH002877.2
Identify how each of F.A.I.R principles has been met.
Alternative records to check: https://www.uniprot.org/uniprot/P42212 https://synbiohub.org/public/bsu/SubtilinReceiver_spaRK_separated/1
Solution
All entries are uniquely identified by a stable URL (‘F’), that provides access to the record in a variety of formats including a web page, plain-text, FASTA, and GENBANK (‘A’, ‘I’). The record contains rich metadata (‘R’) that is both human-readable (HTML) and machine-readable (text) (‘I’). There search options that uses both records IDs and the rich metadata (‘F’). The graphical pane shows how the data are re-usable. Metadata uses ontological terms (e.g. taxonomy) and shared vocabularies (e.g. genbank features) (‘I’). Interlinking with other databases: UniProt, Pubmed (‘I’, ‘R’) enabling automated retrieval of records and cross-referencing information.
FAIR and You (5)
The FAIR acronym is sometimes accompanied with the following labels:
- Findable - Citable
- Accessible - Trackable and countable
- Interoperable - Intelligible
- Reusable - Reproducible
Using those labels as hints discuss how FAIR principles directly benefit you as the data creators.
Solution
- Findable data have their own identity, so they can be easily cited and secure the credits to the authors
- Data accessibility over the Internet using standard protocols can be easily monitored (for example using Google analytics). This results in metrics on data popularity or even geo-locations of data users.
- Interoperable data can benefit the future you, for example you will be able to still read your data even when you no longer have access to the specialized, vendor specific software with which you worked with them before. Also the future you may not remember abreviations and ad-hoc conventions you used before (Intelligible).
- Well documented data should contain all the details necessary to reproduce the experiments, helping the future you or someone taking over from you in the laboratory.
FAIR vs Open Science
FAIR does not mean Open. Actually, FAIR guideline only requires that the metadata record is always accessible. For example, the existence of the data can be known (their metadata), the data can have easy to use PID to reference them, but the actual data files can only be downloaded after the login and authorization.
However, if data are already in the FAIR form, i.e. accessible over the internet, in interoperable format and well documented, then it is almost effortless to “open” the dataset and make it available to the whole public. The data owner can do it any time when he no longer perceives oppening as a risk.
At the same time, Open data which does not follow FAIR guidelines have little value. If they are not well described, not in open formats then they are not going to be re-used even if they were made “open” by posting them on some website.
FAIR Quiz (2 … run through break)
Which of the following statements is true/false (T or F).
F in FAIR stands for free. F
Even if there is no license information you can combine data with yours to produce a plot as long as you give appropriate credit to the other authors. F
Only the figure presenting results of statistical analysis need numerical data that were used to create that figure. F
Sharing numerical data as a .pdf in Zenodo is FAIR. F
Sharing numerical data as an Excel file via Github is not FAIR. F*
Metadata standards (for example MIAME MIQUE) assure the “IR” in FAIR. T
Group websites are one of the best places to share your data. F
Data from failed experiments are not re-usable. F
Data should always be converted to Excel files or .cvs in order to be FAIR. F
A DOI of a dataset helps in getting credit. T
FAIR data are peer reviewed. F
FAIR data have to accompany a publication. F
Solution
copied form above once the wording is corrected
Key Points
FAIR stands for Findable Accessible Interoperable Reusable
FAIR assures easy reuse of data underlying scientific findings
Introduction to metadata
Overview
Teaching: 10 min
Exercises: 20 minQuestions
What is metadata?
What do we use metadata for?
Objectives
Recognise what metadata is
Distinguish different types of metadata
Understand what makes metadata interoperable
Know how to decide what to include in metadata
What is (or are) metadata?
Simply put, metadata is the data about the data. Does this sound confusing? Let’s clarify: metadata is the description of your data. It allows others to gain deeper understanding about your data and provides insight for its interpretation. Hence, you should consider your metadata as important as your data. Further, metadata plays a very important role in making your data FAIR. It has to be continuously added to your research data (not just at the beginning or end of your project!). Metadata can be produced in an automated way (e.g.: when you create a microscopy image usually the accompanying software saves metadata on it) or manually.
Let’s take a look at an example:
This is a confocal microscopy image of a C. elegans nematode strain used as a proteostasis model (Pretty! Isn’t it?). The image is part of the raw data associated to Goya et al., 2020, which was deposited in a Public Omero Server.
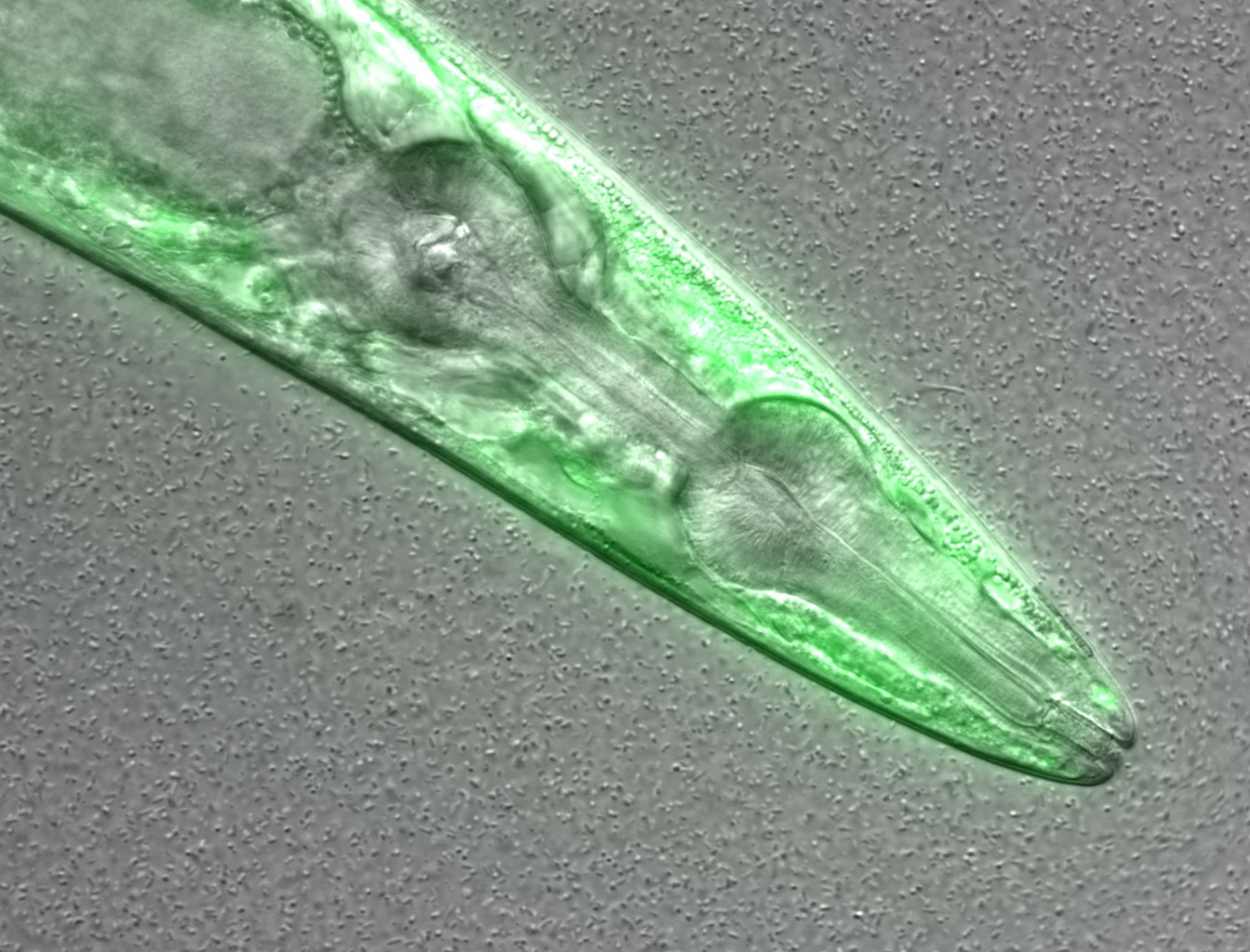 Figure credits: María Eugenia Goya
Figure credits: María Eugenia Goya
. What information can you guess without the associated description (metadata)?
Let’s see the associated metadata to the image and the dataset to which it belongs:
Image metadata
Name: OP50 D10Ad_06.czi Image ID: 3485 Owner: Maria Eugenia Goya ORCID: 0000-0002-5031-2470
Acquisition Date: 2018-12-12 17:53:55 Import Date: 2020-04-30 22:38:59 Dimensions (XY): 1344 x 1024 Pixels Type: uint16 Pixels Size (XYZ) (µm): 0.16 x 0.16 x 1.00 Z-sections/Timepoints: 56 x 1 Channels: TL DIC, TagYFP ROI Count: 0
Tags: time course; day 10; adults; food switching; E. coli OP50; NL5901; C. elegans
Dataset metadata
Name: Figure2_Figure2B Dataset ID: 263 Owner: Maria Eugenia Goya ORCID: 0000-0002-5031-2470
Description: The datasets contains a time course of α-syn aggregation in NL5901 C. elegans worms after a food switch at the L4 stage:
E. coli OP50 to OP50 Day 01 adults Day 03 adults Day 05 adults Day 07 adults Day 10 adults Day 13 adults
E. coli OP50 to B. subtilis PXN21 Day 01 adults Day 03 adults Day 05 adults Day 07 adults Day 10 adults Day 13 adults
Images were taken at 6 developmental timepoints (D1Ad, D3Ad, D5Ad, D7Ad, D10Ad, D13Ad)
* Some images contain more than one nematode.
Each image contains ~30 (or more) Z-sections, 1 µmeters apart. The TagYFP channel is used to follow the alpha-synuclein particles. The TL DIC channel is used to image the whole nematode head.
These images were used to construct Figure 2B of the Cell Reports paper (https://doi.org/10.1016/j.celrep.2019.12.078).
Creation date: 2020-04-30 22:16:39
Tags: protein aggregation; time course; E. coli OP50 to B. subtilis PXN21; food switching; E. coli OP50; 10.1016/j.celrep.2019.12.078; NL5901; C. elegans
This is a lot of information!
Types of metadata
According to How to FAIR we can distinguish between three main types of metadata:
- Administrative metadata: are data about a project or resource that are relevant for managing it; for example, project/ resource owner, principal investigator, project collaborators, funder, project period, etc. They are usually assigned to the data, before you collect or create them.
- Descriptive or citation metadata: are data about a dataset or resource that allow people to discover and identify it; for example, authors, title, abstract, keywords, persistent identifier, related publications, etc.
- Structural metadata: are data about how a dataset or resource came about, but also how it is internally structured. Structural metadata describe, for example, the unit of analysis, collection method, sampling procedure, sample size, categories, variables, etc. Structural metadata have to be gathered by the researchers according to best practice in their research community and will be published together with the data.
Descriptive and structural metadata should be added continuously throughout the project.
Where does data end and metadata starts?
What is “data” and what is “metadata” is can be a matter of perspective: Some researchers’ metadata can be other researchers’ data.
For example, a funding body is a typical administrative metadata, however, it can be used to calculate numbers of public datasets per funder. And then used to compare effect of different funders’ policies on open practices.
Identifying metadata types (3+2 minutes)
Here we have an excel spreadsheet that contains project metadata for a made-up experiment of plant metabolites
Figure credits: Tomasz Zielinski and Andrés Romanowski
In groups, identify different types of metadata (administrative, descriptive, structural) present in this example.
Solution
- Administrative metadata marked in blue
- Descriptive metadata marked in orange
- Structural metadata marked in green
Figure credits: Tomasz Zielinski and Andrés Romanowski
Being precise
If the metadata purpose is to help understand the data, it has to be done in a precise and “understandable” way i.e. it has to be interoperable. To be interoperable metadata should use a formal, accessible, shared, and broadly applicable terms/language for knowledge representation.
One of the easiest examples is the problem of author disambiguation.

After Libarary Carpentry FAIR Data
Open Researcher and Contributor ID (ORCID)
Have you ever done a search in pubmed and found that you have doppelganger? So how can you uniquely associate something you created to just you and no other researcher that has the same name?
ORCID iD is a free, unique, persistent identifier that you own and control—forever. It distinguishes you from every other researcher across disciplines, borders, and time.
ORCIDs of authors of this episode are:
You can connect your iD with your professional information—affiliations, grants, publications, peer review, and more. You can use your iD to share your information with other systems, ensuring you get recognition for all your contributions, saving you time and hassle, and reducing the risk of errors.
If you do not have an ORCID, you should register to get one!
ORCID provides the registry of researchers, so they can be precisely identified. Similarly, there are other registries that can be used to identify many of biological concepts and entities:
- species e.g. NCBI taxonomy
- chemicals e.g. ChEBI
- proteins e.g. UniProt
- genes e.g. GenBank
- metabolic reactions, enzymes e.g KEGG
NCBI or BioPortal are good places to start searching for a registry or a term.
Public ID in action (3)
Wellcome Open Research journal uses ORCID to identify authors.
- Open one of our papers doi.org/10.12688/wellcomeopenres.15341.2 and check how public IDs as ORCID can be used to interlink information.
- The second metadata example (the Excel table) contains two other types of public IDs. Can you find them? Can you find the meaning behind those Ids?
If you have not done it yet, register yourself at ORCID
Solution
ORCID is used to link to authors profiles which list their other publications.
The metadata example contains genes IDs from The Arabidopsis Information Resource TAIR and metabolites IDs from KEGG
Adding metadata to your experiments
Good metadata are crucial for assuring re-usability of your outcomes. Adding metadata is also very time-consuming process if done manually, so collecting metadata should be done incrementally during your experiment.
As we saw metadata can take many forms from as simple as including a ReadMe.txt file, by embedding them inside the Excel files, to using domain specific metadata standard and format.
But,
- what to include in the metadata?
- what public IDs to use?
For many assay methods and experiment types, there are defined recomendations and guidelines called Minimal Information Standards.
Minimal Information Standard
The minimum information standard is a set of guidelines for reporting data derived by relevant methods in biosciences. If followed, it ensures that the data can be easily verified, analysed and clearly interpreted by the wider scientific community. Keeping with these recommendations also facilitates the foundation of structuralized databases, public repositories and development of data analysis tools. The individual minimum information standards are brought by the communities of cross-disciplinary specialists focused on the problematic of the specific method used in experimental biology.
Minimum Information for Biological and Biomedical Investigations (MIBBI) is the collection of the most known standards.
FAIRSharing offers excellent search service for finding standards
What can you do if there are no metadata standards defined for your data / field of research?
Think about the minimum information that someone else (from your lab or from any other lab in the world) would need to know about your dataset to be able to work with it without any further inputs from you.
Think as a consumer of your data not the producer!
What to include - discussion (5+5 minutes)
Think of the data you generate in your projects, and imagine you are going to share them.
What information would another researcher need to understand or reproduce your data (the structural metadata)?
For example, we believe that any dataset should have:
- a name/title
- its purpose or experimental hypothesis
Write down and compare your proposals, can we find some common elements?
Solution
Some typical elements are:
- biological material, e.g. Spciecies, Genotypes, Tissues type, Age, Health conditions
- biological context, e.g. speciment growth, entrainment, samples prepartions
- experimental factors and conditions, e.g. drug treatments, stress factors
- specifics of data aquisition
- specifics of data processing and analysis
Metadata and FAIR guidelines
Metadata provides extreme valuable information for us and others to be able to interpret, process, reuse and reproduce the research data it accompanies.
Because metadata are data about data, all of the FAIR principles i.e. Findable, Accessible, Interoperable and Reusable apply to metadata.
Ideally, metadata should not only be machine-readable, but also interoperable so that they can interlink or reasoned about by computer systems.
Attribution
Content of this episode was adapted from:
Key Points
Metadata provides contextual information so that other people can understand the data.
Metadata is key for data reuse and complying with FAIR guidelines.
Metadata should be added incrementally throught out the project
It's all about planning
Overview
Teaching: 10 min
Exercises: 12 minQuestions
What is the research data life cycle?
How to plan for FAIR sharing?
What is a Data Management Plan (DMP)?
Objectives
Learn what actions can help in achieving FAIR.
Learn to plan for different stages/steps of the data life cycle.
Draft a simple DMP for your project.
The research data life cycle
The Research Data Life Cycle is composed of a sequential series of stages/steps in which data is produced, processed and shared. The arrows between circles represent the transitions that occur in research as work is finished and passed to the next stage/step. The re-use is the driving force of the cycle, cause as if you (or others) were not going to use the data you would not need to store it or even process it.
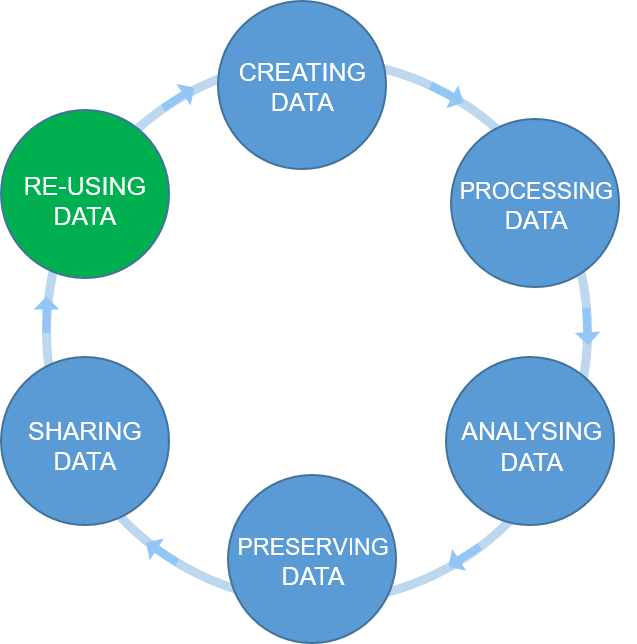 Figure credits: Tomasz Zielinski
Figure credits: Tomasz Zielinski
Data management is a continuous process
Data management should be done throughout the duration of your project. If you wait till the end, it will take a massive effort on your side and will be more of a burden than a benefit.
There are many actions/steps which you can take during your research workflow which would make it easier to share your data in a Findable, Accessible, Interoperable and Reusable way, while, helping you in your day to day activities.
Action plan challenge (4 + 4 minutes)
Where would you say the following actions belong in the Research Data Life Cycle? How do they help in achieving FAIR principles?
- clarify usage rights
- give credit through citations
- use open source software
- attach PID to your data
- attach descriptive metadata
- produce standard metadata
- backup your data
- create figures and plots in python/R
- organize your files in folders
- select data repository
- add open licence
- link publications, data and methods
- create a template for assay description
- use institutional repositories
- use controlled vocabularies
- convert numerical data to csv
- track versions of files
- performing statistical analysis
- deposit datasets to Zenodo/Dryad
- record experiment details in Electronic Lab Notebook
- use github for your code
- ask someone to revise your project structure
- reformat and clean data tables
- use a Minimal Information Standard
- use PID in data description
- download a dataset
- link to UniProt or GenBank records
Solution
Actually many of the above steps can be taken throughout the data life cycle.
Plan ahead: data management plans (DMPs)
A good data management is about PLANning!
The (Data Management Plan) DMP’s purpose is to make you think about your data before you even start the experiments. It should show that you are thinking about what will happen with your data during and after the project. Which of the actions mentioned above you will take and how you will execute them. Finally, how you are going to achieve FAIR.
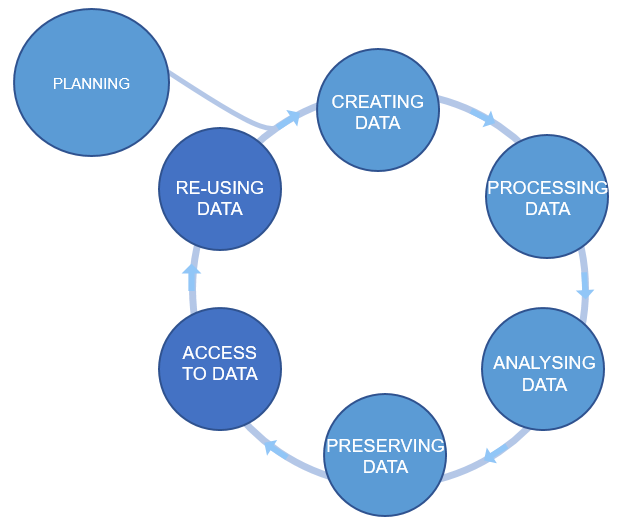 Figure credits: Tomasz Zieliński and Andrés Romanowski
Figure credits: Tomasz Zieliński and Andrés Romanowski
You should think about:
- how you will store the data
- how you will organize and describe your data
- how you will grant access to your data
- how you will share your data
- how you will preserve your data
- how others can use your data
- how much it will all cost
Most of the funders require that you present a DMP together with your grant applications. Some institutions ask eve their PhD students to prepare a DMP for their PhD project.
You should think how you are going to manage your data (our outputs, in general) for each of your projects (or even individual assay types). For individual project the main focus should be on: what data will be produced, how they will be stored and organized, how you are going to describe them and track them. For example what file formats will be generated, how you are going to name your files, and how you will link it to your laboratory notes.
For grant applications, DMP tend to be less technical, for example no need to discuss folder structures, but, they should emphasize the data safety (as preservation and access), data longevity, sharing, discovery and re-use.
DMP Online
DMP Online is a UK tool that is available via subscription to many UK Universities and Institutions. It contains DMP templates for the different funders requirements and information on how to fill each section. Whenever you need to write DMP for a grant application, check if this resource is available to you.
Additionally, your own Institutions may have resources to help with your DMP.
Authors of this course, have created ready to use paragraphs for writing DMPs and a list of recommended repositories at BioRDM wiki
Challenge (given as homework but starts in class 5 minutes)
Working in pairs, think of your last paper (or project). Pretend that you have a joined project that combines the outputs of both your papers/projects.
Write a short DMP for this joined project.
Your DMP should contain the following three sections:
What data you will acquire during the project. Please describe the type of data you will generate (for example ‘flow cytometry data’) as well as file formats and data volume. these data will be stored under (include the meta data as well). Estimate the size of your data.
How you will store the data Please describe how you will store and organize your data, what metadata will you capture in a what form. Tell how you or document the data during the duration of the project
How you will share the data Please describe the strategies for data sharing, licensing and access information.
Remember it is a joined project
Solution
An example DMP can look like:
(1) The project will generate a combination of qualitative and quantitative data derived from phenotyping, LC/MS metabolomics, and general molecular biology techniques.
The main data types and their formats are:
- Phenotyping images (tiff)
- Time-series / Numerical data (Excel)
- MS Metabolomics (mzML)
- processing scripts (Python, R)
The project will generate a total of 4 Tb data.
(2) The instrument specific raw data will be converted into the open formats mentioned above. Daily experimental work will be recorded using an electronic lab notebook (Benchling). We will use ISA templates from MetaboLights for MS data.
All the research data will be stored using the University file system. This is a high quality storage with guaranteed backup. Scripts/codes will be stored under version control using GitHun.
(3) Metabolomics data will be made available through MetaboLights (https://www.ebi.ac.uk/metabolights/) repository. The remaining datasets will be made available through Zenodo. The data will be released no later than a year after the project ends. All data will be made available under CC-BY and the code under MIT licensing.
Attribution
Content of this episode was adapted after:
Key Points
Data within a project go through a set of steps known as the research data life cycle.
Planning can help make your data FAIR.
Data management is a continuous process during a project.
DMP is the best way to prepare for new project.
Record keeping
Overview
Teaching: 20 min
Exercises: 40 minQuestions
How and why to keep good quality records for experiments?
How to streamline and simplify record keeping and its maintenance with electronic lab notebooks and online protocols?
How to keep records FAIR?
Objectives
Use Benchling to create an electronic lab notebook entry with electronic protocols
Reuse and modify an existing lab notebook entry, and export the modified version
Use an existing protocol on protocols.io and adapt it to your own needs
Find out where to track the history of a record in Benchling and protocols.io
Before we start…
Before we start this session on good record keeping, it might be a good idea to make ourselves cup of tea. Here’s a peer-reviewed protocol for making tea:
 Figure credits: Ines Boehm and Ben Thomas
Figure credits: Ines Boehm and Ben Thomas
Differences between analog and record keeping
How did you find making your cup of tea from the above protocol? As you could see, one scientist put milk in before boiling water, another one put boiling water in before milk. Another couldn’t find which type of milk was used. Whilst the steps can be followed to produce a cup of tea, the teas can vary wildly. Slight variations are acceptable, but such as in an experiment, it is more important that steps can be repeated correctly so that others can do the same.
Here is the same protocol typed out on protocol.io. Which do you find easier to follow?
Although digital protocols are the better of the two, analogue protocols certainly have their place. For example, it is often easier to make notes on analogue protocols which you can return to later. Additionally, your working environment may make it too risky to bring expensive technology (just in case), whereas pen and paper can be fine. In cases like these, a hybrid system is often best - where analogue notes are digitized. This can be the best of both worlds.
Challenge (5 minutes)
What are advantages and disadvantages of traditional analog records vs. digital records? Try to find at least a handful of advantages and disadvantages for each. With all of these, which system do you think is most advantageous?
Solution
Advantages of traditional analog records
- Ability to directly draw on your records
- works regardless of internet/power access
Disadvantages of traditional analog records
- can be lost and/or damaged (not Findable or Accessible)
- only in one location at any time (not Findable or Accessible)
- handwriting can make it less intelligible
- harder to edit/move elements around smoothly (not Interoperable)
- can’t store most data types (e.g. imaging data) in a useable way (not Reusable)
Advantages of digital records
- Intelligible: can smoothly and easily move elements around to edit it
- Findable and Accessible: can be shared instantly anywhere around the world, with anyone
- Interoperable: can be easily commented on by anyone anywhere
- doesn’t take up physical space (no record rooms/folders)
- regular backups mean it won’t be lost
- Reusable: version controls mean changes can easily be tracked
- Reusable: can store protocols directly with other supporting data types (e.g. video explanations)
- can you think of more?
Disadvantages of digital records
- dependent on internet access and power (not Accessible)
- some digital record keeping services charge a fee
- risk of corruption if data is not backed up (either yourself or by the service used - not Reusable)
Why do we need to keep good quality records?
Good scientific practice includes good record keeping, which ensures not only transparency and reproducibility, but also accountability. One prime example of why this is necessary is the recent data scandal surrounding Novartis’ FDA approved gene therapy Zolgensma, for the fatal childhood motor neuron disease Spinal Muscular Atrophy (the most expensive treatment ever approved). Novartis submitted manipulated data, showing that the comparison of two versions of Zolgensma in Phase 1 and Phase 3 testing had similar therapeutic activity. How can we prevent the occurrence of data manipulation such as this in the future, or how can we as a research community implement practices to make it easier to find manipulated records? FAIR record keeping, for example version control can help, as it shows what changes have been made when in electronic laboratory notebooks (ELNs), which will make it difficult to manipulate results such as this without leaving a trace.
In order to avoid data mismanagement and such unexplained discrepancies, it is imperative to keep dated, accurate, complete and intelligible records of our experiments and the protocols we use. This means they should include enough detail for others to reproduce under ideally the same conditions. You are, legally (!!) the one responsible for your records, not your colleague, or your PI.
What designates a good record?
Both protocols and laboratory records need to be detailed and kept accurate and complete. They should be accessible (physically and/or electronically) to others both short and long term. Regular back-ups on a cloud and physical hard-drive are necessary to ensure appropriate archiving. All your records should be kept in compliance with departmental, institutional, and other regulatory requirements, with special care given to human and animal research records. A few common guidelines of good record keeping for protocols and laboratory notebooks are the following:
Protocols
- who thought of the protocol if not you
- complete and detailed instructions describing why and how to do an experiment
- what special materials and instruments are being used and where they were obtained
- health and safety advice and how to dispose of waste
- allow repetition of your procedures and studies by yourself and others
Laboratory Notebooks
- contain all relevant details (what, when why and how you did it)
- who you are (the person creating the record)
- what project(s) is the record part of
- information on LOT numbers and batch numbers of cells and antibodies used
- what happened and what did not happen (data, including images)
- how you manipulated and analysed the results
- your interpretation (and the interpretations of others if important) and next steps in the project based on these results
- should be well organised for ease of navigation (indexed, labelled, catalogued)
- accurate and complete: include all irginal data and important study details (metadata) and successful and unsuccessful studies
Why do we want to keep FAIR records?
Focusing on FAIR this keeps our records Findable, Accessible, Interoperable/Intelligible as well as Re-usable. Accessibility that allows better reuse increases our citations and visibility in the field, digital record keeping increases the legibility of notes and provenance (tracking of dates and origins of work) allow for better reproducibility which we have discussed in the previous lesson. Additionally, greater accessibility affords accountability to the original creator of the work. We will now show you how easy it is to share records once they are online, and address some benefits that new repositories such as electronic lab notebooks (ELNs) or online protocols have. There are multiple repo’s for ELNs and online protocols. We will discuss two free options that are easy to use: Benchling and procols.io If you have not created accounts yet for both of them, please do so now as you will need them for the following exercises.
How records become FAIR
Electronic lab notebooks
By now you should be familiar with the concept of why digital record keeping is important, as this is ultimately more reproducible. It is self-explanatory that handwritten notes and laboratory notebook entries take more time to search when looking for an entry. Electronic lab notebooks allow organisation by project, and the search function, or filters, can quickly find what we are looking for. We will now show how to reuse a lab entry that has already been created by someone else.
ELN excercise (5 minutes):
To highlight how easy it is to reuse protocols someone else has used in their lab entry, integrate these into your electronic lab notebooks, and export these for e.g. printing we will be looking at the following Benchling lab entry, making Breakfast. ~~~
Re-use a published lab entry
- First within your own workspace click the big ‘+’ (Create Project) right next to Projects in your Benchling workspace
- Call the project ‘Breakfast’, and add an appropriate description, click ‘Create project’
- Click on the above ‘Benchling lab entry’ link bringing you to the public lab entry ‘Eggs Florentine in Portobello Mushrooms’.
- Select the clock symbol on the right hand side underneath Share: Now you can see the history of the entry and changes that have previously been made to the document with a timestamp. If someone had tried to ‘manipulate’ data, you would be able to see this here. You also see the owner of the document.
- Click ‘Clone from version’
- Select the ‘Breakfast’ folder to clone it to
With your newly set up lab entry, play around with it to explore the interface. Add or remove some text, use the tool to embed a picture etc… You can add text beside the image to ‘annotate’ this appropriately for example. Explore the various things you can interact with to get an understanding of the interface on Benchling.
Adapting a protocol to your needs (6 minutes)
- You have now accessed a digital record and want to reuse it to make your own breakfast. To show how reusable digital recors are we will first navigate through the cloned file you made in your project.
- Navigate to your Project ‘Breakfast’, you can tell you are in your Project, if your initials show in a red circle next to entries in the side bar. You should see the lab entry ‘Eggs Florentine in Portobello Mushrooms’, and the top bar above the title and toolbar should read ‘Tea’, ‘Portobello Mushrooms and Spinach’, ‘Poached Egg and Hollandaise Sauce’, ‘Add Protocol’, ‘Notes’, and ‘Metadata’.
- Click through those tabs and you will see that in your notes you have your lab entry describing how breakfast was made with embedded graphics and a shopping list and current prices. The other three tabs describe the protocols that were used, and you can add additional protocols with the ‘add protocol’ tab. We want you to adapt the ‘Tea’ protocol to suit your ingredients and methods.
- Once you have made appropriate changes in the Tea protocol, you should consider changing the order in which the breakfast and tea are made.
- Once you have made all suggested change have a look at the history of the record (clock button), you can see the changes you have recently made, and you can see it still relates to the original document. It tells you what record it has been cloned from and when.
- Click the link to the original record. As you can see digital record keeping allows provenance, crediting the original author, but also allowing you to keep track of your sources.
- Navigate back to your lab entry in your project (your initials are a sign that you are in the right place).
You hopefully should have set your tea making protocol up so that it’s just the way you like it. Moreover, it should have been easy to make the changes to the protocol. Looking at the history of the record allows you to see the original protocol from which you adapted, and linked your adaptation.
How easy it is to share your record (4 minutes)
- Click the info icon on the right hand side underneath the clock symbol you used previously and select ‘Export entry’
- Your export is now running, you will receive an email when the export is complete
- Click the link in the email to download your protocol as a .zip
- Unzip the file and in your own time, print the protocol if you want to use the recipe in the kitchen, or share it with friends.
- You can share .pdf versions, or click Share and generate a Share link of your lab entry. This makes your record interoperable as many users across many platforms across the world can access your entry if you make it public and share it on for example social media. If there is no IT access present, you always have the option to print the .pdf copy. ~~~
Now that you have your export you can easily share it with others and use it yourself elsewhere, while your digital record (and links to the original record) are maintained online. You can share the PDF that you have exported with others (or print it and store it the lab/workspace), or link them to your digital record, allowing them to make further changes if you wish.
Electronic Protocols
As you could see, Benchling has an integrated platform for protocols. But there are other repositories, such as protocols.io that we briefly mentioned beforehand, which have been developed to help make protocols FAIR. One can publish protocols with a DOI which has many benefits that we discussed in the previous lesson. Another strength is, that you can create a protocol, fork the original protocol if your lab started implementing small changes, and retain both versions that you can cite in your publications or share with your collaborators.
Adapt public protocols and retain provenance (10 minutes)
To copy protocols from protocols.io, edit them and export them as a .pdf there are a few simple steps to follow. We will be using the making a cup of tea protocol.
# Fork the protocol, preserving the original for crediting 1. Open the link to the above protocol, as you can see we have assigned it its own DOI 2. First click on Metrics: Because we are FAIR, this shows you how how many views over time this protocol has had, how many exports, how many individual steps it involves and how many times it has been forked. 3. Now click on the downwards arrow next to the title 4. Select 'Copy/Fork' and click 'make a fork' 5. Select the Folder you want the protocol to be forked to and click 'continue' 6. Your fork of "How to make a cup of tea" is ready now, click 'edit new fork' 7. On the right hand tool bar, the clock icon, shows you the history of the protocol (as before in Benchling). Currently you should see no history as you have not made changes. # Edit the forked protocol 1. Go to 'Materials' in the top tool-bar: add or edit materials according to your preferences, e.g. change full-fat milk to oat-milk, or add honey, lemon etc 2. Go to 'Steps' in the top tool-bar: edit the protocol according to your preferences 3. You can edit the 'Description' and 'Guidelines & Warnings' if you would like to 4. As soon as you change anything, the timestamp and where in the protocol this change was made appears in the history. 5. Click 'View', you will now see the reader view of your protocol. It clearly states underneath the title 'Forked from How to make a cup of tea' and the original protocol is linked. This allows clear identification of your source. 6. Click 'Edit' # Optional: Export the forked protocol 1. Click 'More' in the top tool-bar, select 'Export' > 'PDF' > 'To your computer' and click export (leave selections blank)
Now, if you go back to the original protocols.io protocol that you have forked from, and click on metrics, you will see how that views have increased. Additionally, if you click on ‘Forks’, it will show you that there are a multitude of private forks and if you click on one of these forks, e.g. ‘Cup of tea (polish way)’ you will be taken to the protocol. A particular perk of this is accessibility and accompanying accountability to the original creator of the work.
How to choose the right platform?
There are more than a 100 platforms that provide services to host electronic lab notebooks or protocols, therefore it can seem quite daunting trying to find the right platform. There is some advice we can offer when looking for the right service.
- Make sure they are in compliance with departmental, institutional, and other regulatory and legal requirements (including where data is geographically located, and which types of data can and cannot be stored).
- You want an acceptable pricing model (is it free?), check if your institution has a subscription to any.
- What is the longevity of the ELN (is it a brand new ELN, or has it been established for a while now, i.e. is there a risk of the ELN folding & data being lost?).
- What is the ability to share & export entries, experience of colleagues, availability of support, integration with other relevant platforms (e.g. dropbox), and the potential for use with mobile devices if required?
- What is the user interface like? Does it feel intuitive or does it take you days to find what you are looking for.
- Check for operating system compatibility and real time collaboration.
- Preferably it should be Open Source.
The BioRDM team has put together a comprehensive summary of ELNs on the University of Edinburgh Wiki where they test-ran a handful of ELNs for you so you can make a more informed choice.
It is always best to use a free version or trial version to test an ELN first and see which features you are missing and which ones you prefer. Some often used ones are:
- Scinote (free version available)
- Rspace (trial possible)
- Benchling (free version available)
- Labarchives (trial possible contacting the company)
- WikiBench (works on top of Confluence)
ELN challenge (7 minutes)
Do you use an ELNs? Which one? What features do you like?
How does good record keeping help us get FAIR ready?
Accessibility of your protocols online allows to share them with collaborators who need them for a publication and a DOI of said protocols allows to cite them with credibility. Electronic lab records allow for easier re-usability and access of your data across multiple platforms. Changes in your records can be traced back, therefore giving accountability.
FAIR record keeping - Quiz (3 minutes)
Which of the following statement are true/false.
- Good record keeping ensures transparency and reproducibility. (T)
- There are no advantages to using analog record keeping when compared to digital record keeping. (F)
- Digital records help people view a protocol simultaneously. (T)
- Digitally kept records can be quickly and easily edited. (T)
- On balance, digital record keeping is more advantageous than analog record keeping. (T)
- Digital records are easier to search than analog records. (T)
Solution
- Good record keeping ensures transparency and reproducibility. (T)
- There are no advantages to using analog record keeping when compared to digital record keeping. (F)
- Digital records help people view a protocol simultaneously. (T)
- Digitally kept records can be quickly and easily edited. (T)
- On balance, digital record keeping is more advantageous than analog record keeping. (T)
- Digital records are easier to search than analog records. (T)
Further Reading
Key Points
Good record keeping ensures transparency and reproducibility.
Record keeping is an integral part of data FAIRification.
Record keeping is key to good data management practices.
Working with files
Overview
Teaching: 15 min
Exercises: 16 minQuestions
How should I name my files?
How does folder organisation help me
Objectives
Understand elements of good naming strategy
Evaluate pros and cons of different project organizations
Explain how files management helps in being FAIR
Project organization: planning file names and folders structure
Before you even start collecting or working with data, you should decide how you will structure and name files and folders. This will:
- allow for standardized data collecting and analysis by many team members.
- make it easier for the researcher to determine where files should be saved.
- avoid file duplication.
- help to make retrieval and archiving more efficient.
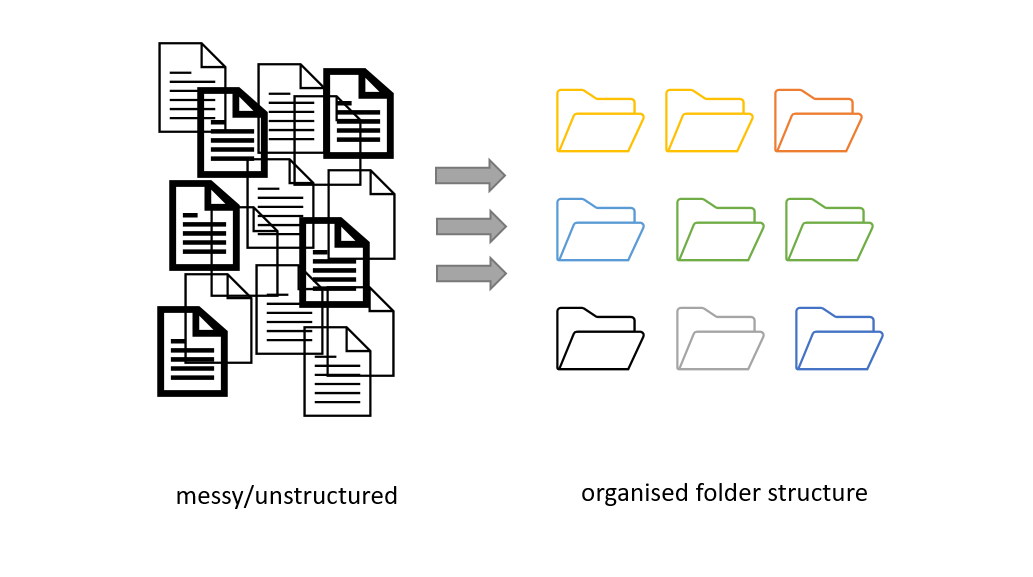 Figure credits: Andrés Romanowski
Figure credits: Andrés Romanowski
Consistent naming and organizing files in folders has two main goals:
- quick finding needed files
- being able to tell the file content without opening it
Naming your files (and folders)
One important and often overlooked aspect of organizing, sharing, and keeping track of data
files is standardising naming.
It is important to develop naming conventions
which permits encoding experimental factors which are important to the project.
File (folder) names should be consistent, meaningful to you and your collaborators, allow you to easily find what you are looking for, give you a sense of the content without opening the file, and identify if something is missing.
Naming and sorting (3+2 minutes)
Have a look at the example files from a project, similar to the one from metadata episode.
All the files have been sorted by name and demonstrate consequences of different naming strategies.
For your information, to encode experimental details, following conventions were taken:
- phyB/phyA are sample genotype,
- sXX is sample number
- LD/SD are different light conditions (long or short day)
- on/off are different media (on sucrose, off sucrose)
- measurement date
- other details are timepoint and raw or normalized data
2020-07-14_s12_phyB_on_SD_t04.raw.xlsx
2020-07-14_s1_phyA_on_LD_t05.raw.xlsx
2020-07-14_s2_phyB_on_SD_t11.raw.xlsx
2020-08-12_s03_phyA_on_LD_t03.raw.xlsx
2020-08-12_s12_phyB_on_LD_t01.raw.xlsx
2020-08-13_s01_phyB_on_SD_t02.raw.xlsx
2020-7-12_s2_phyB_on_SD_t01.raw.xlsx
AUG-13_phyB_on_LD_s1_t11.raw.xlsx
JUL-31_phyB_on_LD_s1_t03.raw.xlsxLD_phyA_off_t04_2020-08-12.norm.xlsx
LD_phyA_on_t04_2020-07-14.norm.xlsx
LD_phyB_off_t04_2020-08-12.norm.xlsx
LD_phyB_on_t04_2020-07-14.norm.xlsx
SD_phyB_off_t04_2020-08-13.norm.xlsx
SD_phyB_on_t04_2020-07-12.norm.xlsx
SD_phya_off_t04_2020-08-13.norm.xlsx
SD_phya_ons_t04_2020-07-12.norm.xlsx
ld_phyA_ons_t04_2020-08-12.norm.xlsx
- What are the problems with having date first?
- How do different date formats behave once sorted?
- Can you tell the importance of leading 0 (zeros)?
- Is it equally easy to find all data from LD conditions as ON media?
- Can you spot problem with when using different cases?
- Do you see benefits of keeping consistent lengths of each name parts?
- Do you see what happens when you mix conventions?
Solution
- Using dates up front makes it difficult to quickly find data for particular conditions or genotypes. It also masks the “logical” order of samples or timepoints.
- Named months break the “expected” sorting, same as dates without leading 0
- Without leading zeros, ‘s12’ appear before s1 and s2
- the first (and second) parts of the name are easiest to spot
- last file is also from LD conditions but do apearch after SD, same with ‘phya’ genotypes
- the last 3 file names are easiest to read as all parts appear on top of each other, thanks to using same 3 letter-lemgth codes ons and off
- The lack of consistency makes it very difficult to get data from related samples/conditions.
Some things to take into account to decide on your naming convention are:
- Does your convention make your files easy to sort
and find by the most important feature?
- include any parameter that helps the name being as descriptive as possible (i.e.: project, experiment, researcher, sample, organism, date (range), data type, method)
- defined a standard vocabulary (shortcuts) for parameters
- decide which elements go in which order.
-
Decide the convention when to use symbols, capitals, hyphens (e.g kebab-case, CamelCase, or snake_case).
-
Defined a maximum name length. Aim for filenames no longer than ~30 characters.
- Document any abbreviation of your parameters.
Do’s:
- for dates use the YYYY-MM-DD standard and place at the end of the file UNLESS you need to organise your files chronologically
- include version number (if applicable), use leading zeroes (i.e.: v005 instead of v5).
- make sure the 3-letter file format extension is present at the end of the name (e.g. .doc, .xls, .mov, .tif)
- add a PROJECT_STRUCTURE (README) file in your top directory which details your naming convention, directory structure and abbreviations
Don’ts:
- avoid using spaces (use _ or - instead)
- avoid dots, commas and special characters (e.g. ~ ! @ # $ % ^ & * ( ) ` ; < > ? , [ ] { } ‘ “)
- avoid using language specific characters (e.g óężé), unfortunately they still cause problems with may software or between operating systems (OS)
- avoid long names
- avoid repetition, e.g if directory name is Electron_Microscopy_Images, and file ELN_MI_IMG_20200101.img then ELN_MI_IMG is redundant
- avoid deep paths with long names (i.e. deeply nested folders with long names) as archiving or moving between OS may fail
If adding all the relevant details to file names makes them too long, it is often a signal that you should use folder to organize the files and capture some of those parameters.
Folders vs Files (3 minutes)
Have a look as these two different organization strategies:
(1) |– Project
|– |– arab_LD_phyA_off_t04_2020-08-12.metab.xlsx(2) |– Project
|– |– arabidopsis
|– |– |– long_day
|– |– |– |– phyA
|– |– |– |– |– off_sucrose_2020-08-12
|– |– |– |– |– |– t04.metab.xlsxCan you think of scenarios in which one is better suited than other? Hint: think of other files that could be present as well.
Solution
The first strategies, can work very well if the project has only few files, so all of them can quickly be accessed (no need to change folders) and the different parameters are easily visible. For example a couple of conditions, couple of genotypes or species
– Project – – arab_LD_phyA_off_t04_2020-08-12.metab.xlsx – – arab_LD_WILD_off_t03_2020-08-11.metab.xlsx – – arab_SD_phyA_off_t01_2020-05-12.metab.xlsx – – arab_SD_WILD_off_t02_2020-05-11.metab.xlsx – – rice_LD_phyA_off_t05_2020-05-02.metab.xlsx – – rice_LD_WILD_off_t06_2020-05-02.metab.xlsx – – rice_SD_phyA_off_t07_2020-06-02.metab.xlsx – – rice_SD_WILD_off_t08_2020-06-02.metab.xlsx The second strategy works better if we have a lot of individual files for each parameter. For example, imagine the metabolites are measured hourly throughout the day, and there are ten different genotypes, two species and 4 light conditions. You would not want to have all the 2000 files in one folder.
– Project – – arabidopsis – – – long_day – – – – phyA – – – – – off_sucrose_2020-08-12 – – – – – – t01.metab.xlsx – – – – – – t02.metab.xlsx – – – – – – t03.metab.xlsx – – – – – – … – – – – – – t23.metab.xlsx – – – – – – t24.metab.xlsx – – rice – – – long_day – – – – phyA – – – – – off_sucrose_2020-06-03 – – – – – – t01.metab.xlsx – – – – – – … – – – – – – t24.metab.xlsx
Must do: Document your strategy
Regardless of whether you are using long filenames or incorporating some of the variables within the folder structure, document it!
Always include a PROJECT_STRUCTURE (or README) file describing your file naming and folder organisation conventions.
Strategies to set up a clear folder structure
Establishing a system that allows you to access your files, avoid duplication and ensure that your data can be easily found needs planning.
You can start by developing a logical folder structure. To do so, you need to take into account the following suggestions:
- Use folders to group related files. A single folder will make it easy to locate them.
- Name folders appropriately: use descriptive names after the areas of work to which they relate.
- Structure folders hierarchically: use broader topics for your main folders and increase in specificity as you go down the hierarchy.
- Be consistent: agree on a naming convention from the outset of your research project.
Good enough practices for scientific computing recommendations
The Good enough practices in scientific computing paper makes the following simple recommendations:
- Put each project in its own directory, which is named after the project
- Put text documents associated with the project in the ‘doc’ directory
- Put raw data and metadata in a ‘data’ directory
- Put files generated during cleanup and analysis in a ‘results’ directory
- Put project source code in the ‘src’ directory
- Put compiled programs in the ‘bin’ directory
- Name all files to reflect their content or function:
- Use names such as ‘bird_count_table.csv’, ‘notebook.md’, or ‘summarized_results.csv’.
- Do not use sequential numbers (e.g., result1.csv, result2.csv) or a location in a final manuscript (e.g., fig_3_a.png), since those numbers will almost certainly change as the project evolves.
Organization for computing (3 minutes)
Take a look at the folder structure recommended by the Good enough practices in scientific computing paper.
Why do you think it is recommended layout and suited for a computing project?
.
|– CITATION
|– README
|– LICENSE
|– requirements.txt
|
|– data
| |– birds_count_table.csv
|
|– doc
| |– notebook.md
| |– manuscript.md
| |– changelog.txt
|
|– results
| |– summarized_results.csv
|
|– src
| |– sightings_analysis.py
| |– runall.py
|Solution
This project structure clearly separates the inputs (the raw data) from the outputs (the results) and the analysis procedure (python code). Following the same convention (like src folder for code) makes it easy to find interesting elements, for example the raw data or particular ploting procedure.
The root directory contains a README file that provides an overview of the project as a whole, a CITATION file that explains how to reference it, and a LICENSE, all three make it REUSABLE. The src directory contains a controller script runall.py that loads the data and triggers the whole analysis.
After you have a plan
Your naming conventions might need some adjustments as the project progresses. Don’t despair, just document it!
If you change the strategy, document it in PROJECT_STRUCTURE (or README) stating why you made the change and when. Update the locations and names of files which followed the old convention
Backing up your project files and folders
- Back up (almost) everything created by a human being or recorded by a machine as soon as it is created.
- Always backup your files in 3 places, at least one should be off-site.
- USB sticks are a failure-prone option and are not valid solution for backup of scientific data
- A robust backup cannot be achieved manually
Do you know how and where to keep 3 copies of your data which are always up to date?
Secure data preservation is very difficult to achieve without institutional support and know-how. One option is a cloud storage, but not all data may not be put in a public cloud.
You should always check your institutional guidelines and what solutions are available in your organization.
Project files organization and FAIR guidelines
FAIR Files (3+2 minutes)
In groups, discuss:
- how can strategy for folder organisation and naming convention help in achieving FAIR data?
Have you realised that the following the above suggestions means including valuable metadata as part of your folder structure and file names?
Where to next
Bulk renaming of files can be done with the software such as Ant Renamer, RenameIT or Rename4Mac.
Good enough practices in scientific computing (Wilson et al., 2017)
Attribution
Content of this episode was created using the following references as inspiration:
Key Points
A good file name hints the file content
Good project organization saves you time
Describe your files organization in PROJECT_STRUCTURE
(Meta)data in Excel
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Does and donts in Excel
Data cleaning
Excel vs cvs
How tidy data helps FAIR
As in objectives, ?maybe mention adding PIDs where possible, or discuss how to future proof your data for example have a tab in which shortcuts for strains or conditions are explained then they could be also enriched with PID rather than use PIDs through out documents as those are prone to human errors unlike labels which are easy to spot or correct?
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points
Templates for consistency
Overview
Teaching: 0 min
Exercises: 0 minQuestions
How to achieve consistancy with templates
Use PIDs and ontology terms in RightField template
Define own RightField template
How it helps with FAIR
Showin righfield templates provided they still works and there is nothing better
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points
Jupyter notebooks for data analysis
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Benefits of notebooks for analysis
Use a notebook and modify it
How notebooks help in FAIR
Show a notebook (with nice descriptions cells) which:
- (if possible to get a sensible library/program) calls a code from command line to do some simple file processing (maybe one step from workflows workshop) that should show that it can replace commands lines while showing how exactly the programs/library well called
- reads in the results or excell files (in python)
- plots something from the results
-
saves the figure to a file
- ask to run it
- ask to add some description (non coding cell)
- ask to change input file to another one and re-analysed it
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points
Version control
Overview
Teaching: 0 min
Exercises: 0 minQuestions
why version control in computing
why version control in docs and data
how to do version control
?minimal git?
Jupyter experience to show need for version control. We changed inputs, getting new figures. Maybe we want the previous one.
For docs the document hell. Copy of copy etc. That is great for figures/results. But for docs? Frankly going through text changes in a docs in git is a nightmare. Much easier review-mode of Word with collored changes etc, but, once accepted those are lost.
Can we actually show how git helps? Using the web ui (preferably). The git workflow is over-complicated. So many commands for simple save. Not so from web UI. Needs careful thinking of what is achievable with the audience.
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points
Public repositories
Overview
Teaching: 10 min
Exercises: 20 minQuestions
Where can we deposit and find research datasets?
What are general (research data) repositories?
What are specific (research data) repositories?
How do repositories help make research data FAIR?
Objectives
See the benefits of using research data repositories.
Be able to find a suitable repository.
Be able to differentiate between general and specific repositories.
See how repositories help make research data FAIR.
What are research data repositories?
Research data repositories are online repositories that enable the preservation, curation and publication of research ‘products’. These repositories are mainly used to deposit research ‘data’. However, the scope of the repositories is broader as we can also deposit/publish ‘code’ or ‘protocols’ (as we will see later).
Research outputs should be submitted to discipline/domain-specific repositories whenever it is possible. When such a resource does not exist, data should be submitted to a ‘general’ repository.
Research data repositories are a key resource to help in data FAIRification.
Challenge 1. The general repository (5 minutes).
Have a look at the following data set in Zenodo: link to Zenodo perfect deposit
Discuss: What elements make it FAIR?Solution
The elements that make this deposit FAIR are:
Findable:
- F1. (Meta)data are assigned a globally unique and persistent identifier - YES
- F2. Data are described with rich metadata (defined by R1 below)- YES
- F3. Metadata clearly and explicitly include the identifier of the data they describe - YES
- F4. (Meta)data are registered or indexed in a searchable resource - YES
Accessible:
- A1. (Meta)data are retrievable by their identifier using a standardised communications protocol - YES
- A2. Metadata are accessible, even when the data are no longer available - YES
Interoperable:
- I1. (Meta)data use a formal, accessible, shared, and broadly applicable language for knowledge representation. - YES
- I2. (Meta)data use vocabularies that follow FAIR principles - PARTIALLY
- I3. (Meta)data include qualified references to other (meta)data - YES
Reusable:
- R1. (Meta)data are richly described with a plurality of accurate and relevant attributes - YES
- R1.1. (Meta)data are released with a clear and accessible data usage license - YES
- R1.2. (Meta)data are associated with detailed provenance - YES
- R1.3. (Meta)data meet domain-relevant community standards - YES/PARTIALLY
Challenge 2. Datasets discovery (4 minutes).
Can you easily find similar data sets in Zenodo? Try to find an interesting dataset for you
Hint 1: Verify the completeness/richness of the associated metadata. Is it complete? Are some bits missing?
Hint 2: Does the dataset include a ReadMe.txt file?Solution
Zenodo is a good place to keep your data separate from paper. It gives access to all files, allowing you to cite the data as well (or instead of) the paper.
However, it is not good for disovery, and does not enforce most metadata!
Challenge 3. Domain specific repositories (4 minutes).
Select one of the following repositories based on your expertise/interests:
- Have a look at mRNAseq accession ‘E-MTAB-7933’ in ArrayExpress
- Have a look at microscopy ‘project-1101’ in IDR
- Have a look at the synthethic part record ‘SubtilinReceiver_spaRK_separated’ within the ‘bsu’ collection in SynBioHub
- Have a look at the proteomics record ‘PXD013039’ in PRIDE
- Have a look at the metabolomics record ‘MTBLS2289’ in Metabolights
- Have a look at the scripts deposit ‘RNA-Seq-validation’ in GitHub
Report to the group, what advantages can you see in using a specific repository over a generalist repository like Zenodo.
Solution
Some advantages are:
- The repository is more relevant to your discipline than a generalist one.
- Higher exposure (people looking for those specific types of data will usually first look at the specific repository).
- Higher number of citations (see above).
How do we choose a research data repository?
As a general rule, your research needs to be deposited in discipline/data specific repository. If no specific repository can be found, then you can use a generalist repository. Having said this, there are tons of data repositories to choose from. Choosing one can be time consuming and challenging as well. So how do you go about finding a repository:
- Check the publisher’s / funder’ recommended list of repositories, some of which can be found below:
- Check Fairsharing recommendations
- alternatively, check the Registry of research data repositories - re3data
Challenge 4. Finding a repository (4 minutes).
a) Find a repo for genomics data.
b) Find a repo for microscopy data.
Note to instructor: Fairsharing gives few options, people may give different answer follow up why they selected particular ones.Solution
a) GEO/SRA and ENA/ArrayExpress are good examples. Interestingly these repositories do not issue a DOI.
b) IDR and UoE Public Omero are good examples. Once again, these repositories do not issue a DOI.
A list of UoE BioRDM’s recommended data repositories can be found here.
What comes first? the repository or the metadata?
Finding a repository first may help in deciding what metadata to collect and how!
Extra features
It is also worth considering that some repositories offer extra features, such as running simulations or providing visualisation. For example, FAIRDOMhub can run model simulations and has project structures. Do not forget to take this into account when choosing your repository. Extra features might come in handy.
Can GitHub be cited?
To make your code repositories easier to reference in academic literature, you can create persistent identifiers for them. Particularly, you can use the data archiving tool in Zenodo to archive a GitHub repository and issue a DOI for it.
Evaluating a research data repository
You can evaluate the repositories by following this criteria:
- quality of interaction: is the interaction for purposes of data deposit or reuse efficient, effective and satisfactory for you?
- take-up and impact: what can I put in it? Is anyone else using it? Will others be able to find stuff deposited in it? Is the repository linked to other data repositories so I don’t have to search tehre as well? Can anyone reuse the data? Can others cite the data, and will depositing boost citations to related papers?
- policy and process: does it help you meet community standards of good practice and comply with policies stipulating data deposit?
An interesting take can be found at Peter Murray-Rust’s blog post Criteria for succesful repositories.
Challenge 5, Wrap up discussion (3 minutes).
Discuss the following questions:
- Why is choosing a domain specific repositories over zenodo more FAIR?
- How can selecting a repository for your data as soon as you do an experiment (or even before!) can benefit you research and help your data become FAIR?
- What’s your favourite research data repository? Why?
Attribution
Content of this episode was adapted or inspired by:.
Key Points
Repositories help researchers share their research data.
Some repositories are general and others are more data-type specific.
Repositories are key players in data reuse.
?Putting it all together?
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Making FAIR project in OSF repository
Is there a value there apart from being hands on? In OSF or in FairdomHub? Or maybe half in one half in another and talk of experiences.
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points
Where to next
Overview
Teaching: 0 min
Exercises: 0 minQuestions
Wrapping up
Better DMP
Where to look for help/resources
People could reflect on DMPs from life-cycle, would they change something now.
Ask if there was something missing in the course Ask what they would like to spend more time on
List some resources.
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points
Template
Overview
Teaching: 0 min
Exercises: 0 minQuestions
I am episode intro
I am a section
With a text.

After Figure source
I am a yellow info
And my text.
I am code
I am a problem
Defined here.
Solution
- I am an answer.
- So am I.
Attribution
Content of this episode was adopted after XXX et al. YYY.
Key Points